Research
-
FORCE FIELDS
Force Field Development
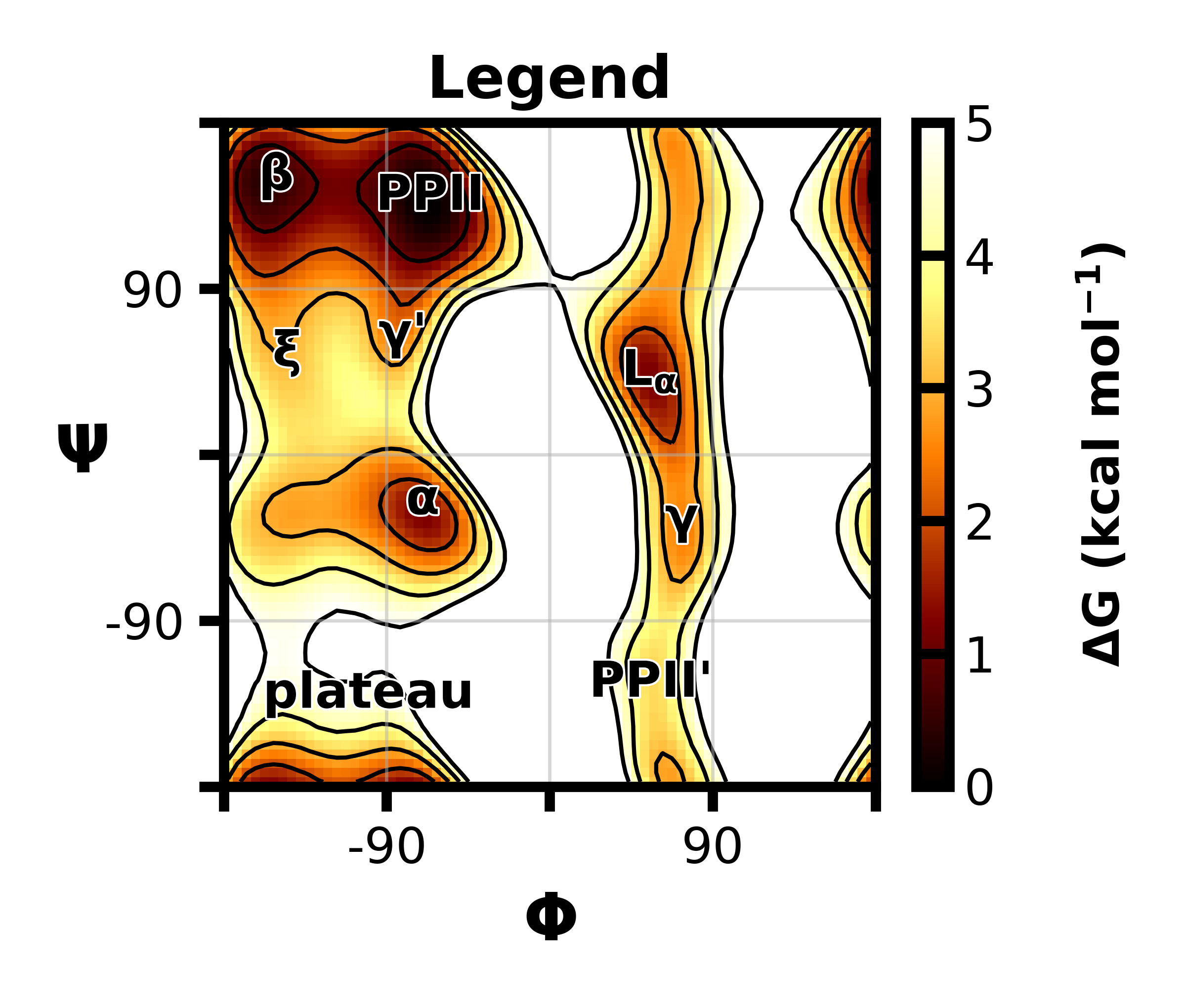 Parameters for the AMBER ff15ipq protein force field. I was a part of the validation efforts for the ff15ipq-m force field [1] and have also fully developed and parameterized ff15ipq compatible parameters for the fluorinated, aromatic amino acids that are commonly used in 19F NMR [2]. To help with validation of the fluorinated amino acid parameters, I developed a way to calculate pragmatically 19F NMR relaxation rates. I've also worked on a more automated workflow for parameterizing new ligand molecules in the context of the ff15ipq force field.
Parameters for the AMBER ff15ipq protein force field. I was a part of the validation efforts for the ff15ipq-m force field [1] and have also fully developed and parameterized ff15ipq compatible parameters for the fluorinated, aromatic amino acids that are commonly used in 19F NMR [2]. To help with validation of the fluorinated amino acid parameters, I developed a way to calculate pragmatically 19F NMR relaxation rates. I've also worked on a more automated workflow for parameterizing new ligand molecules in the context of the ff15ipq force field.
[1] The Journal of Chemical Physics
(2020)
[2] The Journal of Physical Chemistry A(2022)
Code for 19F ff15ipq parameters
Code for general ff15ipq ligand parameterization
Code for calculating 19F NMR relaxation rates from MD simulation data
-
ENHANCED SAMPLING
Rare Event Sampling of Viral Capsids
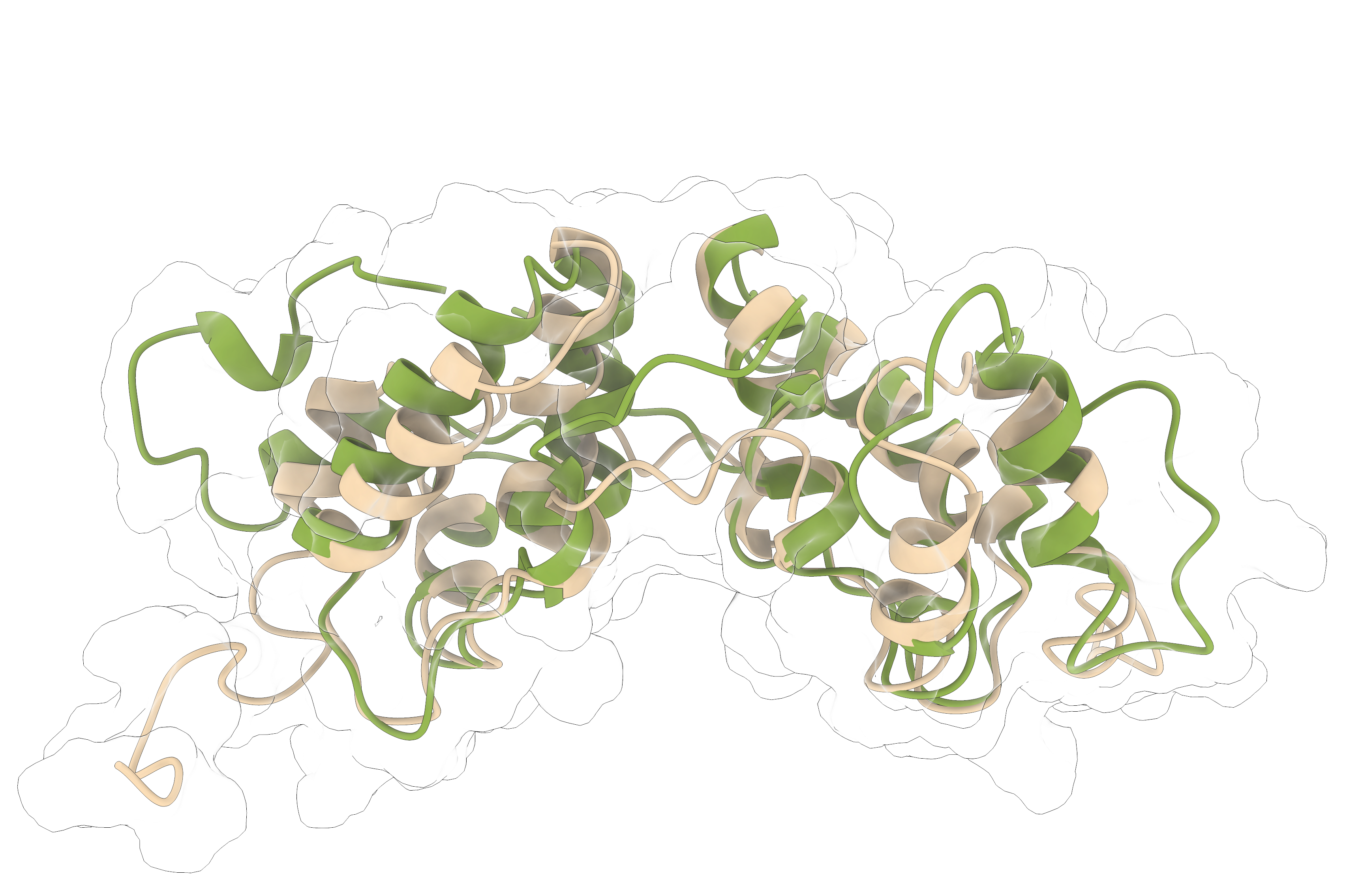 Conformational sampling of the HIV-1 capsid protein dimer using the weighted ensemble path sampling strategy.
Conformational sampling of the HIV-1 capsid protein dimer using the weighted ensemble path sampling strategy.
-
NMR
19F NMR
 By combining a diHis motif, Co2+, and a chelating ligand, we can get orthogonal pseudocontact shift (PCS) datasets. This allows us to look at exact atomic positions within a distance range using the same protein.
By combining a diHis motif, Co2+, and a chelating ligand, we can get orthogonal pseudocontact shift (PCS) datasets. This allows us to look at exact atomic positions within a distance range using the same protein.
[1] The Journal of the American Chemical Society
(2023)
Scripts for MD simulations and QM calculations to support PCS results
-
ANALYSIS TOOLS
Data Analysis and Visualization
 Python package for faster and more accessible analysis of h5 datasets output from WESTPA simulations.
Python package for faster and more accessible analysis of h5 datasets output from WESTPA simulations.